The research area of our lab is cancer systems biology and data mining. For several years, we have developed a series of data mining methods to process biomedical data, aiming to solve the problem of precision medicine. Recently, we are working on cancer early detection by using cell-free DNA fragmentation.
Current research projects
Epigenomics in liquid biopsy for the early diagnosis and prognosis of diseases

The fine-scale cell-free DNA fragmentation patterns in early-stage cancers are poorly understood. We developed a de novo approach to characterize the cell-free DNA fragmentation hotspots from plasma whole-genome sequencing. Hotspots are enriched in open chromatin regions, and, interestingly, 3′end of transposons. Hotspots showed global hypo-fragmentation in early-stage liver cancers and are associated with genes involved in the initiation of hepatocellular carcinoma and associated with cancer stem cells. The hotspots varied across multiple early-stage cancers and demonstrated high performance for the diagnosis and identification of tissue-of-origin in early-stage cancers. We further validated the performance with a small number of independent case–control-matched early-stage cancer samples.
Research on precision medicine based on depth cascade centroid classifier

Precision medicine is to understand and treat diseases by integrating multimodal or multigroup data from individuals, so as to make personalized diagnosis and treatment strategies that match the pathological characteristics of patients. As a complex and diverse disease, tumor has great heterogeneity in molecular genetics. Even cancer patients with the same pathological type may have different responses to anti-cancer drugs, so cancer science has become one of the important fields of precision medicine.
At present, using omics data to make corresponding prediction is a popular solution. However, high feature dimensions and small sample size are common in omics data sets. In this paper, we propose a precision medical research method based on deep cascade centroid classifier, and apply it to early cancer diagnosis, prognosis, drug sensitivity prediction and other fields. Deep cascade centroid classifier is an integrated learning method, which has cascade structure and can perform representation learning.
Systems Pharmacology-Based Precision Therapy and Drug Combination Discovery for Breast Cancer
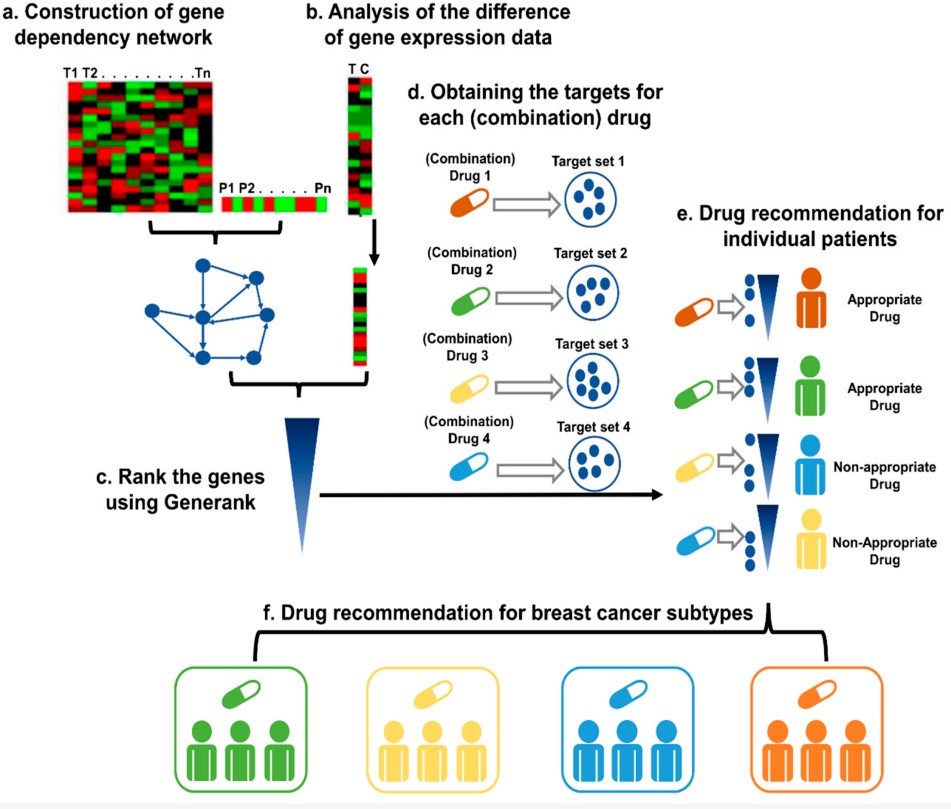
Breast cancer (BC) is a common disease and one of the main causes of death in females worldwide. In the omics era, researchers have used various high-throughput sequencing technologies to accumulate massive amounts of biomedical data and reveal an increasing number of disease-related mutations/genes. It is a major challenge to use these data effectively to find drugs that may protect human health. In this study, we combined the GeneRank algorithm and gene dependency network to propose a precision drug discovery strategy that can recommend drugs for individuals and screen existing drugs that could be used to treat different BC subtypes.
We used this strategy to screen four BC subtype-specific drug combinations and verified the potential activity of combining gefitinib and irinotecan in triple-negative breast cancer (TNBC) through in vivo and in vitro experiments. The results of cell and animal experiments demonstrated that the combination of gefitinib and irinotecan can significantly inhibit the growth of TNBC tumour cells. The results also demonstrated that this systems pharmacology-based precision drug discovery strategy effectively identified important disease-related genes in individuals and special groups, which supports its efficiency, high reliability, and practical application value in drug discovery.