November 1, 2025
Paper published on Briefings in Bioinformatics!

May 1, 2025
Paper published on Cell Reports Methods!
June 24, 2024
Lab Joy by East Lake: An Unforgettable Gathering


We set aside experimental data, relaxing completely amidst the lakeside views: mahjong clashes, patient fishing, board game battles. By evening, we grilled delicious food lakeside—laughter mingling with the sizzling scent. Post-dinner, intense pool showdowns peaked the excitement. Amidst the cheer, our lab bonds deepened!
October 27, 2024
Paper published on Briefings in Bioinformatics!

This study on using multi-omics data for clustering in cancer subtyping is essential for precision medicine. While many methods exist, they often fall short in performance or biological interpretability, hindering practical application. To address this, we developed the Iterative Pathway Fusion approach for enhanced Multi-omics Clustering (IPFMC), which includes two data fusion stages. In the first stage, omics data are partitioned using pathway information, iteratively selecting key pathways to represent samples. In the second stage, similarity network fusion integrates the representation from multiple omics. Comparative experiments with nine cancer datasets from The Cancer Genome Atlas (TCGA) show that IPFMC outperforms ten representative methods. Furthermore, the biological pathways and genes identified by our approach are significant, demonstrating both excellent clustering performance and biological interpretability.
October 22, 2024
Paper published on Clinical Chemistry!

Our results were accepted and published in Clinical Chemistry, this study investigates the potential of cfDNA fragmentomics-based liquid biopsy for noninvasive bladder cancer (BLCA) detection. We assessed the diagnostic performance of cfDNA hotspot-driven machine learning models in a cohort of 55 BLCA patients, 51 individuals with benign conditions, and 11 healthy volunteers. The model demonstrated robust detection capabilities, achieving an area under the curve (AUC) of 0.96, with a sensitivity of 87% and a specificity of 100%. Sensitivity for early-stage and advanced disease was 71% and 92%, respectively. The cfDNA hotspots effectively retrieved regulatory elements and captured molecular features associated with BLCA. These findings support the applicability of urine cfDNA fragmentation hotspots for noninvasive BLCA diagnosis and suggest directions for future research.
June 17, 2024
Paper published: Paper is published on PLOS COMPUTATIONAL BIOLOGY

On August 26, 2024, our research group published a paper titled "IMI-driver: Integrating multi-level gene networks and multi-omics for cancer driver gene identification" in the prestigious international journal PLOS COMPUTATIONAL BIOLOGY. This study innovatively proposes the IMI-driver model, which integrates multi-omics data into eight biological networks and utilizes multi-view collaborative network embedding techniques to successfully compress complex gene regulatory information into a low-dimensional vector space, significantly enhancing prediction accuracy. When tested on 29 cancer types from The Cancer Genome Atlas (TCGA) and multiple benchmark datasets, IMI-driver demonstrated a marked superiority over other methods, further validating its effectiveness. Pan-cancer analysis revealed that the genes identified by this model are predominantly known or potential cancer drivers, and newly discovered positive genes have also been confirmed through case studies to play pivotal roles in cancer progression. This breakthrough not only deepens our understanding of cancer driver mechanisms but also provides robust support for precision medicine strategies in cancer.
June 24, 2024
Grassland Adventure Tale: A Dreamlike Exploration Galloping Through Inner Mongolia




In June 2024, Zhou Lab ushered in the annual carnival tourism season, guess where our destination is, yes, it is Inner Mongolia with a large blue sky and white clouds, green grass! We went to see the volcano together, rode horses and skied grass on the grassland, ran and jumped, sang loudly, ate delicious lamb, drank Mongolia tea, and the whole trip was so happy!
June 24, 2024
Maronghui's Splendid Graduation Season: Setting Sail for Dreams, A Shimmering Farewell Celebration



As Maronghui embarks on a new journey, her splendid graduation season shines brightly, marking the launch of dreams and a dazzling farewell celebration.
June 17, 2024
Paper published: Paper is published on Advanced Science

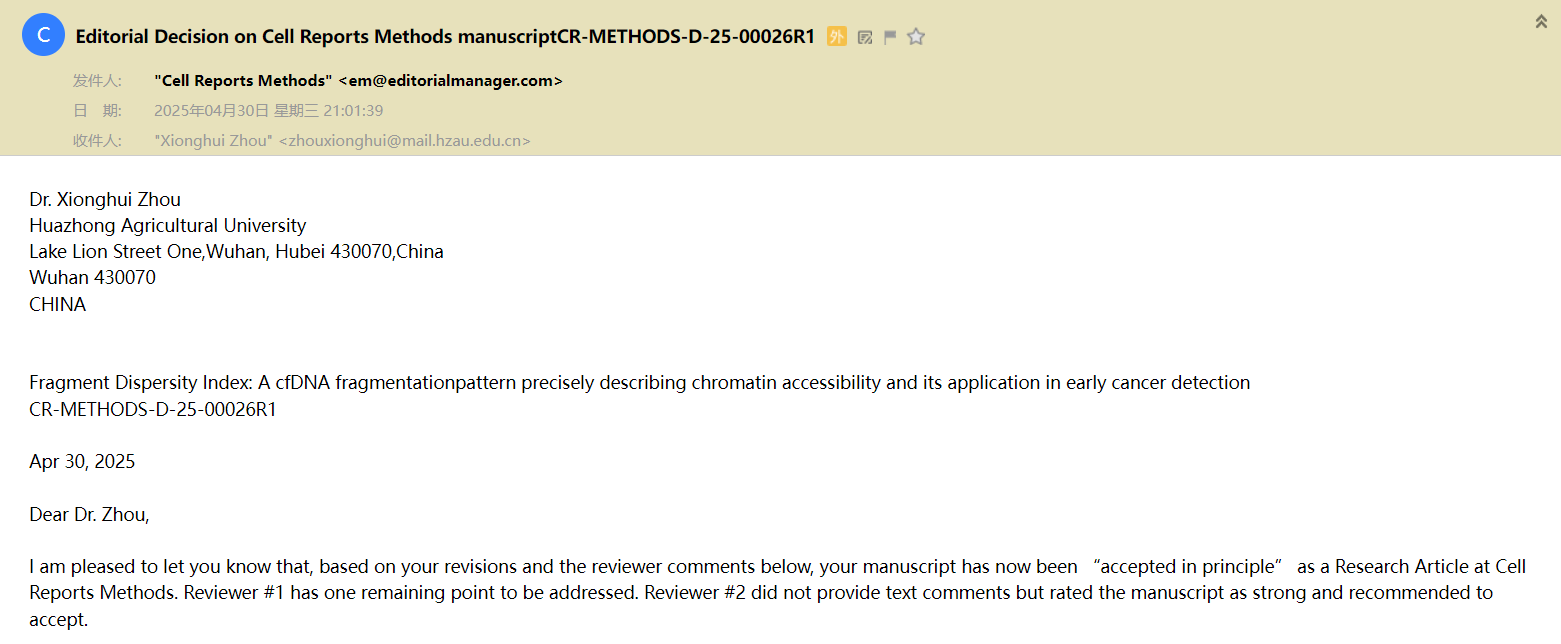
On June 17, 2024, our research group published a paper titled "Systematically Evaluating Cell‐Free DNA Fragmentation Patterns for Cancer Diagnosis and Enhanced Cancer Detection via Integrating Multiple Fragmentation Patterns" in the internationally renowned journal Advanced Science. The main focus of this study is: Cell-free DNA within open chromatin regions is susceptible to fragmentation. This study evaluates the diagnostic performance of 10 representative fragmentation patterns in open chromatin regions across 4 datasets for cancer diagnosis. An ensemble model is constructed by combining these fragmentation patterns, demonstrating a more stable performance for cancer detection and tissue-of-origin determination. The model's crucial regions offer biologically interpretable insights.
February 1, 2024
Paper published: Deep centroid paper is published on Bioinformatics
On February 1, 2024, Contrgulations to Kuan Xie and Yuying Hou, our Deep Cntroid paper has been published on Bioinformatics (Deep centroid: a general deep cascade classifier for biomedical omics data classification).
November 12, 2023
Participated in the 2023 academic Annual meeting of Hubei Key Laboratory of Agricultural Bioinformatics


November 12, 2023 14:30-18:00 PM: Held the 2023 Academic Annual conference of Hubei Key Laboratory of Agricultural Bioinformatics.
Hou Yuying of the research group made a presentation entitled "Systematic Evaluation of Cell-Free DNA Fragmentation Patterns for Cancer
Diagnosis and Enhanced Cancer. Detection through Integration of Multiple Fragmentations "report. The study focused on the use of cell-free
DNA fragments in cancer diagnosis.
The general contents of the study are as follows:
Cell-free DNA (cfDNA) fragmentation patterns hold immense potential for early cancer detection.
However, the definitions of fragmentation vary by genomic scale, ranging from the entire genome to specific genomic regions.
Furthermore, the lack of systematic comparison among these patterns has impeded their broader research and practical implementation.
Here, we collected over 1,382 plasma cfDNA sequencing samples from diverse sources, covering eight cancer types.
Considering that cfDNA within open chromatin regions is more susceptible to fragmentation,
we leveraged ten fragmentation patterns within open chromatin regions as features and employed machine learning techniques to evaluate their performance.
Remarkably, all fragmentation patterns demonstrated discernible classification capabilities,
with EDM (End Motif) showing the highest diagnostic value in cross-validation.
Combining the results of both cross-validation and independent validation,
fragmentation patterns that incorporate both fragment length and coverage information exhibit robust predictive capacities.
Despite their diagnostic potential, these fragmentation patterns exhibited variability in their predictive power.
To address this limitation, we developed an ensemble classifier by integrating all fragmentation patterns,
which achieved significant advancements in cancer detection and tissue-of-origin analysis.
Moreover, thorough biological function investigations of significant feature intervals in our model
have unveiled its impressive ability to discern critical regulatory regions involved in cancer pathogenesis.
June 23, 2023
Drifting with "Roaring in the Sky" and Happy Graduation


Celebrate your graduation journey, enjoy a joyful drifting journey in Yichang, thrilling and thrilling!
May 14, 2023
Home Party: Collision between Delicious and Entertainment

The Data Mining and Precision Medical Laboratory, along with a large group of teachers, launched a Home Party! At Carlton Manor in Optics Valley Square, there are three chefs who offer delicious dishes such as mushroom stewed chicken, minced chili fish, winter yin gong soup, and a large number of dumpling experts, Dragon Flying Phoenix Dance. After the enjoyable food segment, we began a carnival of various games such as mahjong, KTV, billiards, barbecue, computers, and more. After the final entertainment, students need to work harder!
December 8, 2022
Paper published: CRAG paper is published on Genome Medicine

On December 8, 2022, our CRAG paper is published on Genome Medicine (CRAG: De novo characterization of cell-free DNA fragmentation hotspots in plasma whole-genome sequencing). Congratulations!
November 28, 2022
SRF project was awarded the "Top Ten Projects"!
On November 28, 2022, Peiting Shi et al.’s SRF project (Peiting Shi, Junmin Han, Yinhao Zhang and Guanpu Li. IMI-driver, integrating multi-omics for cancer driver identification) was awarded the "Top Ten Projects" in the 2022 Innovation and Entrepreneurship Training Plan of Huazhong Agricultural University. Congratulations!
November 12, 2022
Sports activities: Members of the research group compete passionately through table tennis

On the evening of November 12, 2022, the weather is cold and windy. The four young generals in the laboratory met on the table tennis court to show their skills. To express the pressure of learning in sports, I still don't forget to discuss algorithm ideas.
October 27, 2022
Academic report: Dr. Xiangyu Meng made a speech on "Molecular biology and technology of bladdercancer: APOBEC, tobaco, and generic variability"
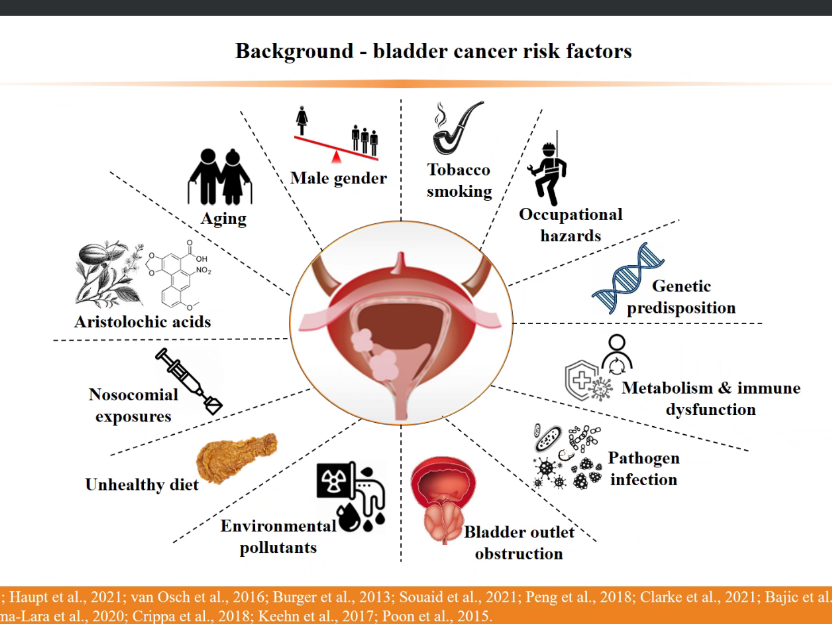
In the afternoon of October 27, 2022, the 18th "Happy Hour" activity of the School of Information in 2022 was held online. At the invitation of Zhou Xionghui, a teacher from the Institute of Information Technology, Dr. Xiangyu Meng made an academic report.
Bladder cancer is the cancer with the highest incidence rate in China, and its clinical treatment has great differences. There are many factors that can induce cancer, such as men's incidence rate may be higher than women's due to smoking, or age may lead to increased cancer risk and occupational exposure. Dr. Meng Xiangyu introduced the statistical analysis of somatic mutation law of bladder cancer and the systematic exploration and integration of genetic variation information of bladder cancer, and then introduced the impact on bladder cancer mutation from three aspects: APOBEC, smoking and genetic variation.
June 10, 2022
Academic report: Xionghui Zhou delivered a speech of "Precision Medical Research Based on System Biology"

Dr. Xionghui Zhou introduced the work of the laboratory in precision medicine. The report mainly started with network construction methods such as "ceRNA network" and "gene dependent network", and then started with "early diagnosis and prognosis of cancer", "drug response prediction", "combination drug prediction" and other aspects.
Precision medicine is a kind of medicine that uses genetic information of individual diseases to guide their diagnosis or treatment. Therefore, based on reasonable biological assumptions, a biological network that can reflect some biological phenomena can be built, and key molecular characteristics of diseases can be mined for early diagnosis, prognosis and drug efficacy prediction.